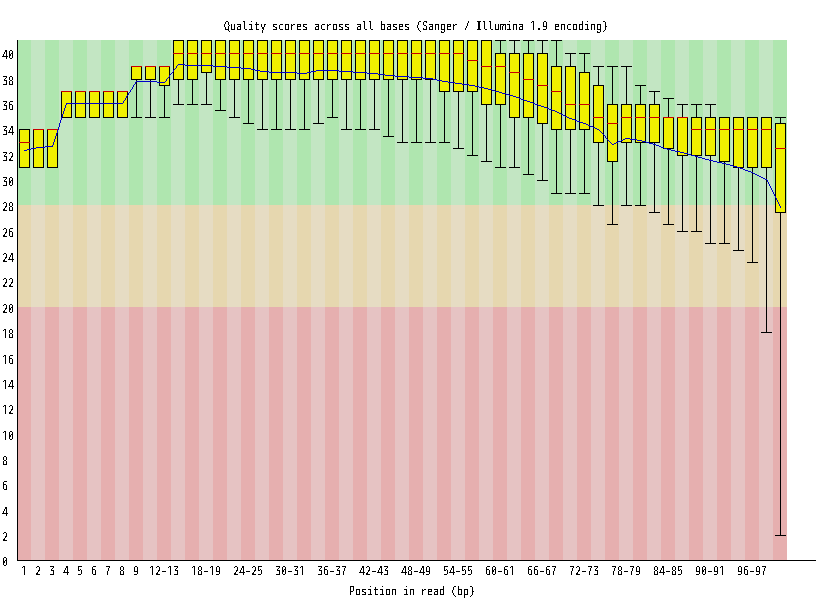
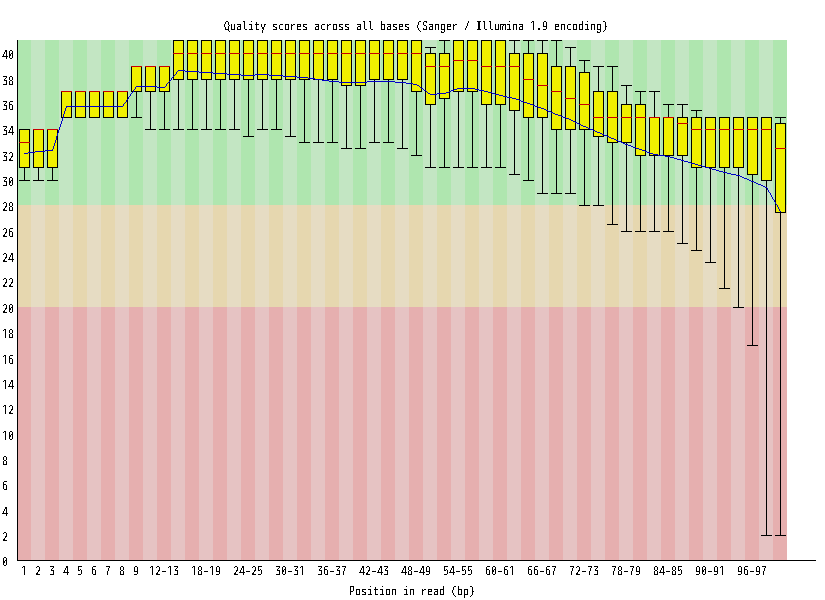
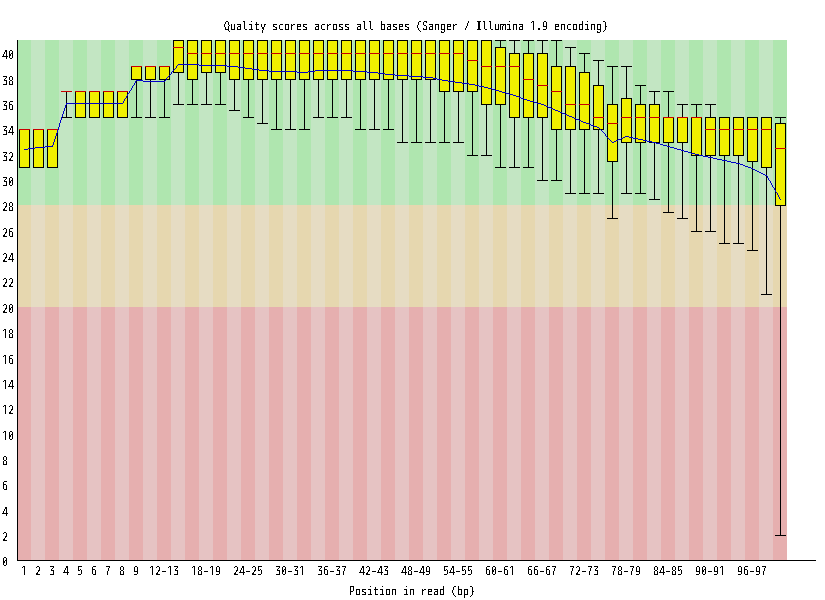
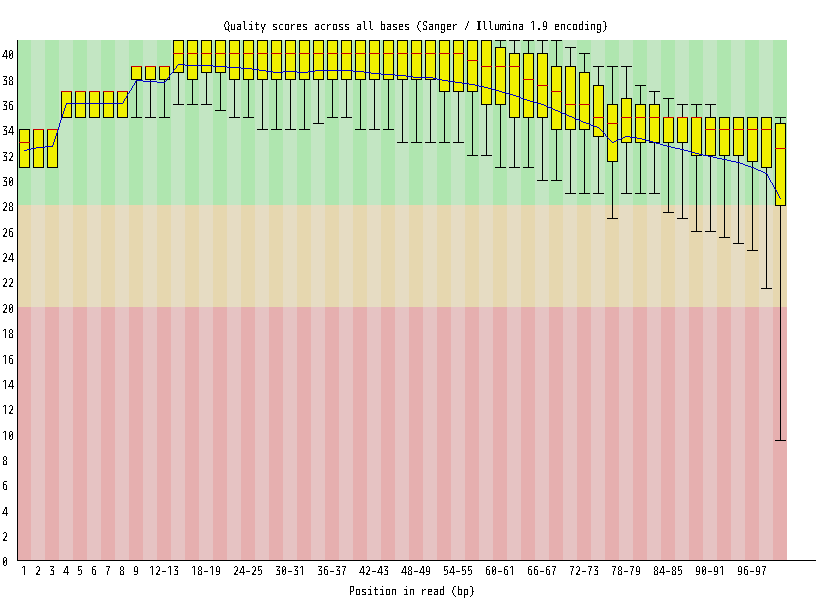
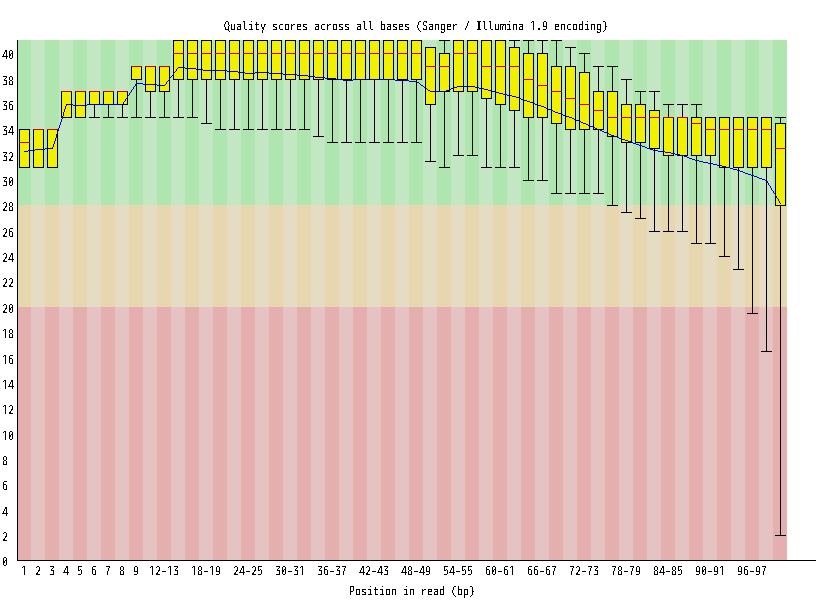
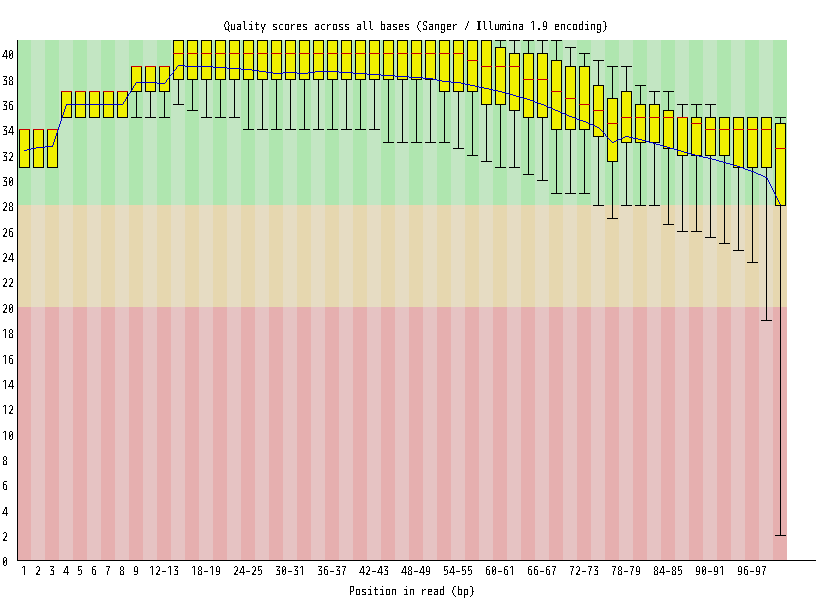
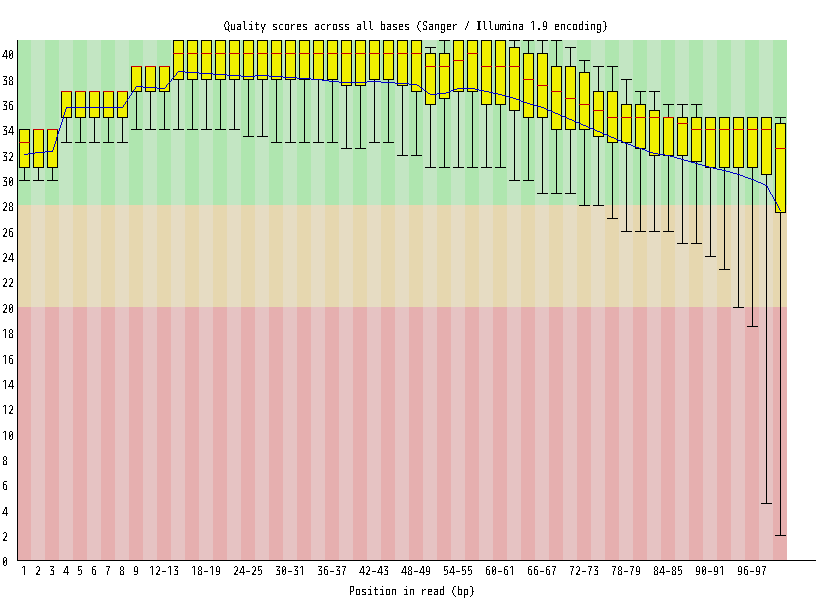
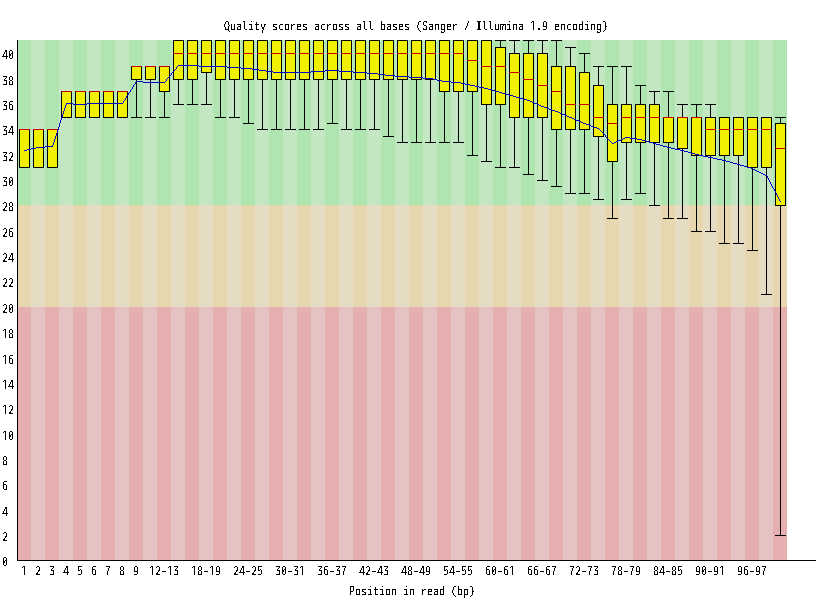
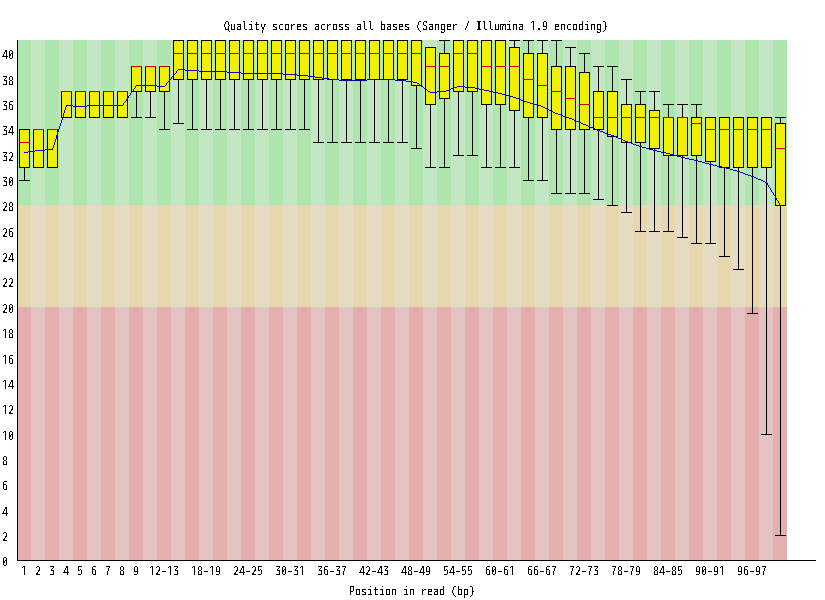
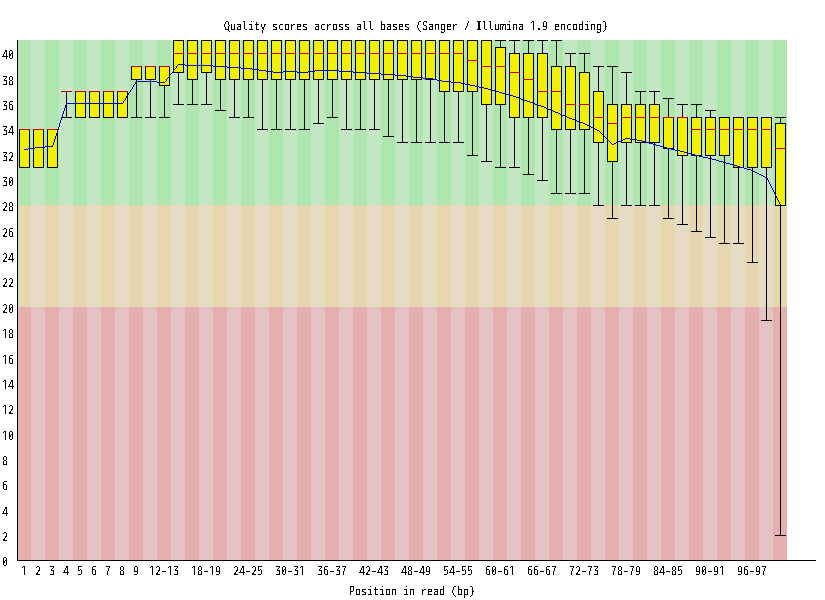
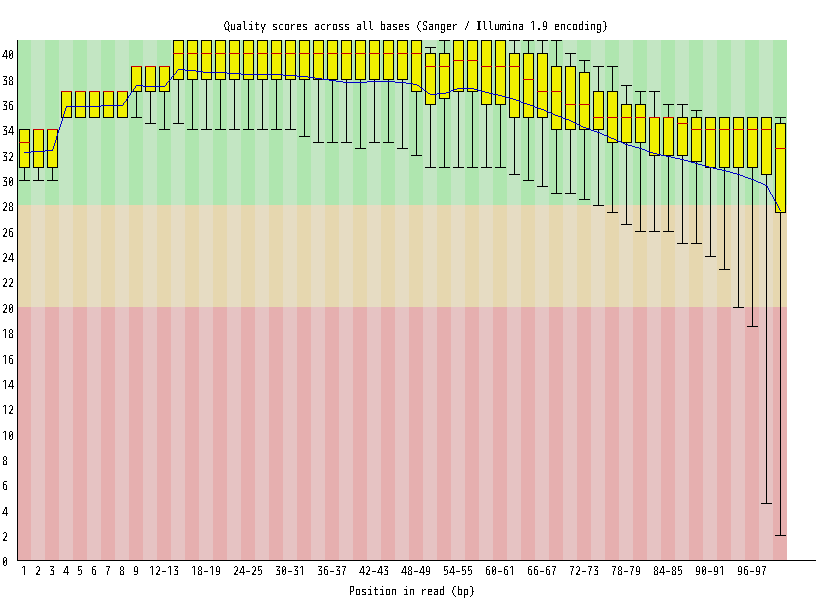
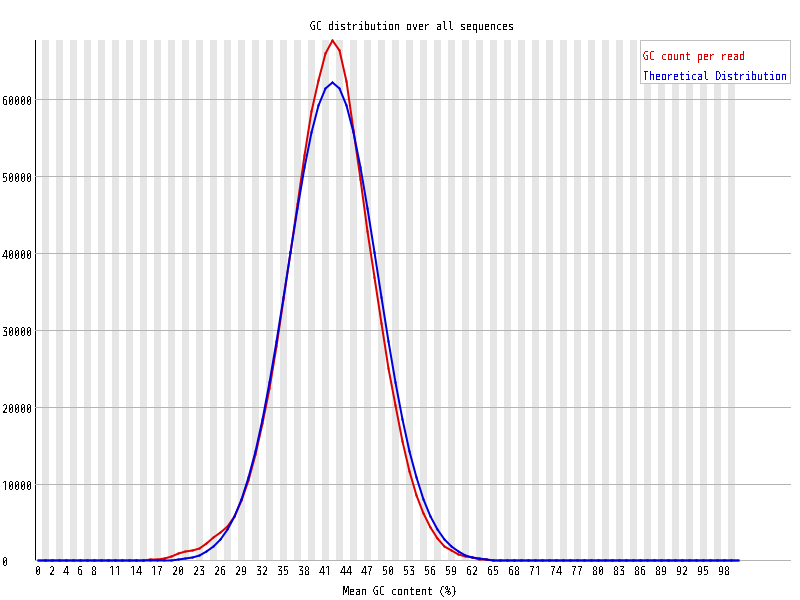
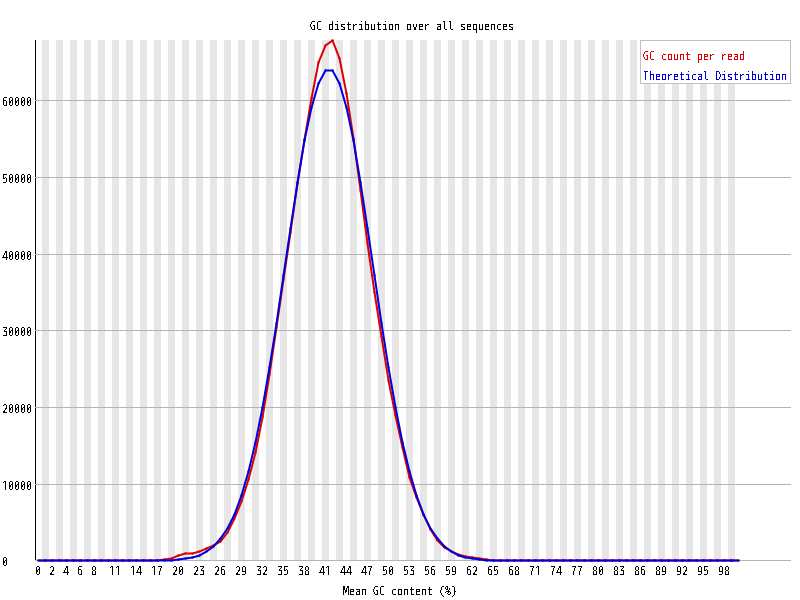
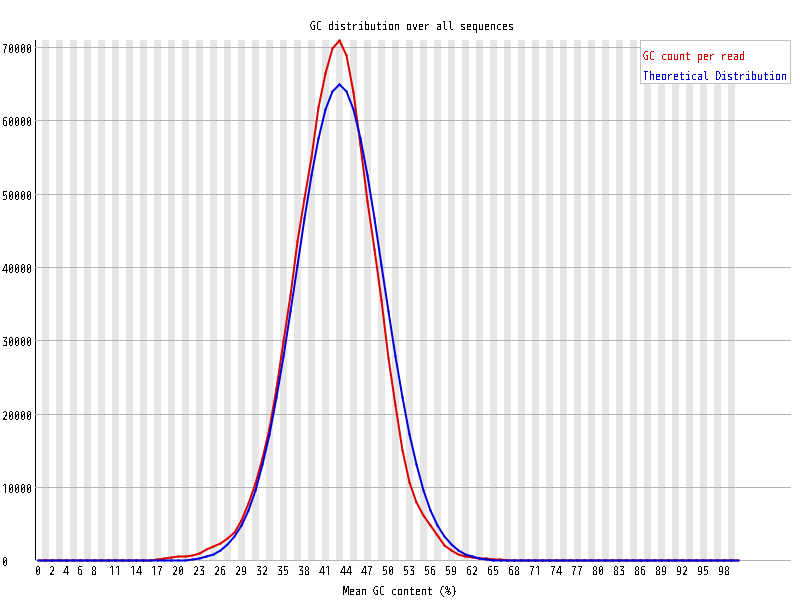
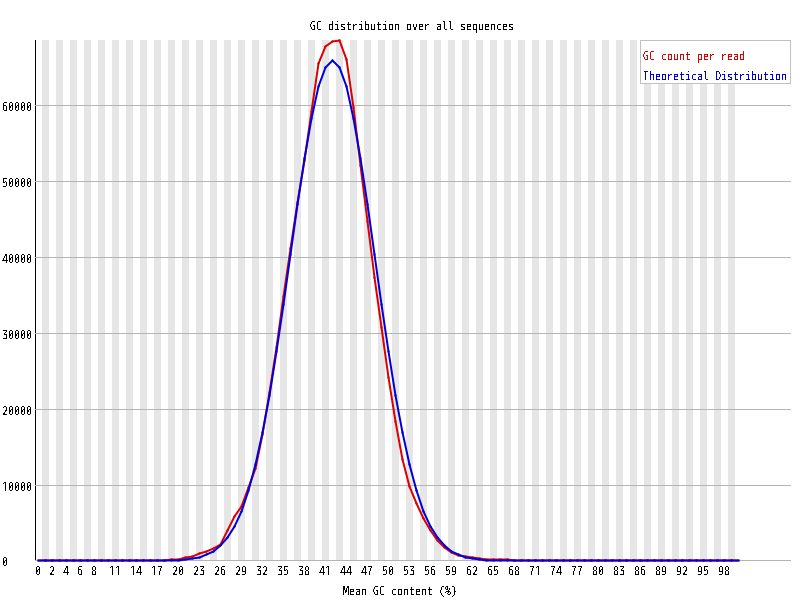
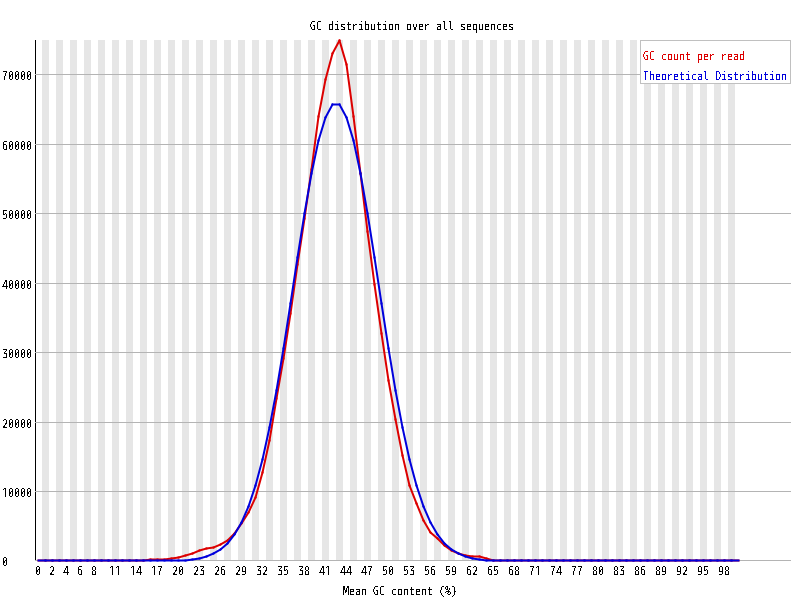
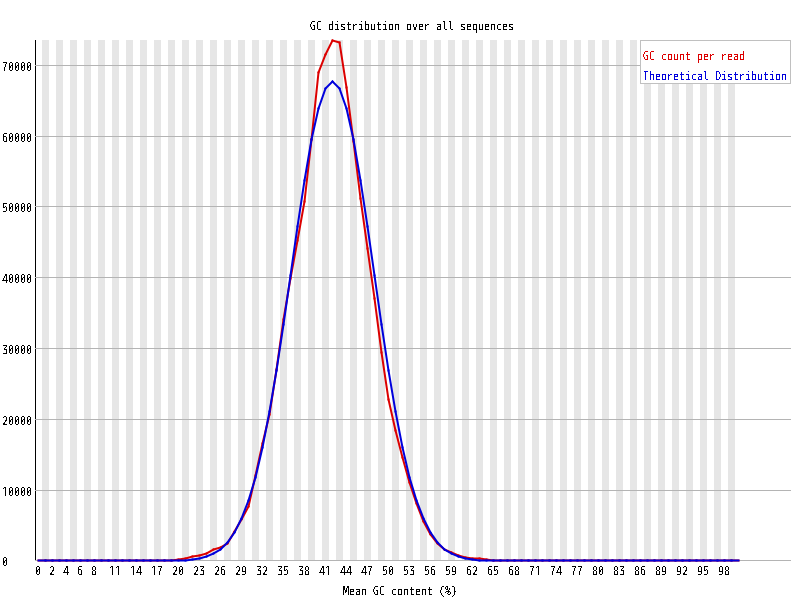
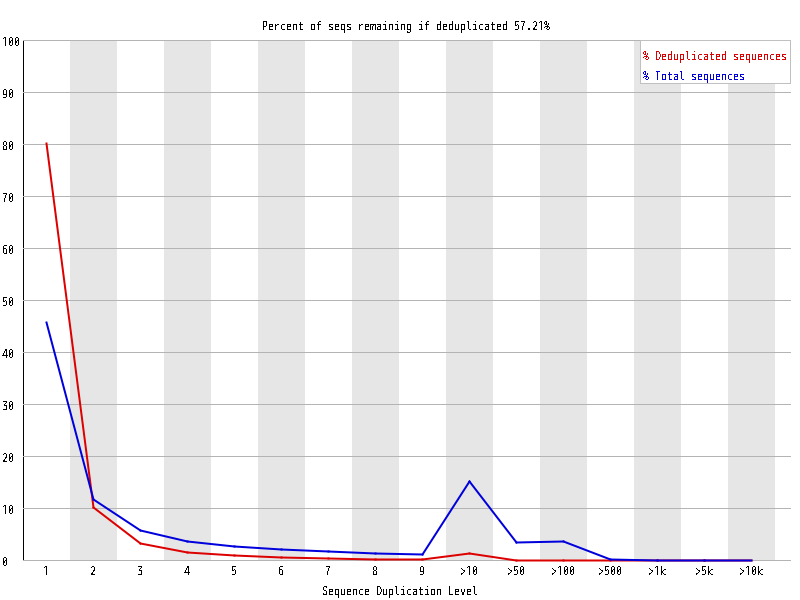
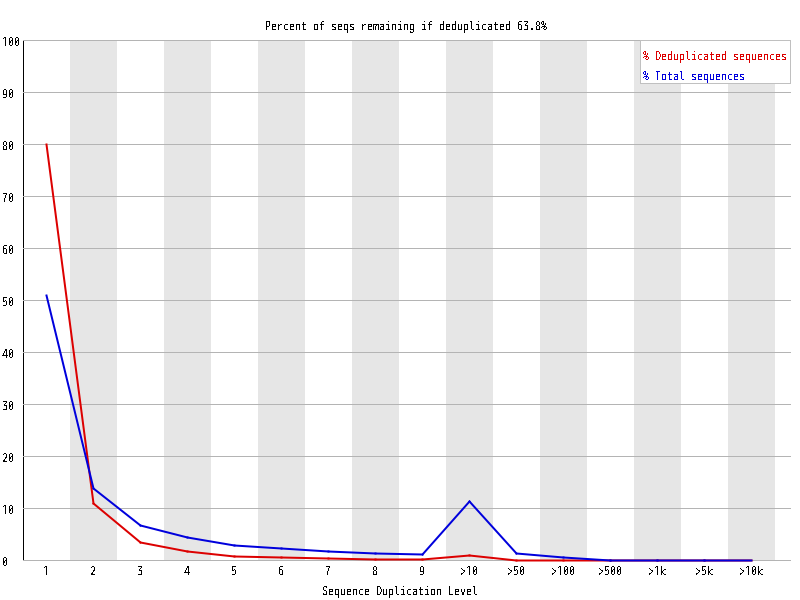
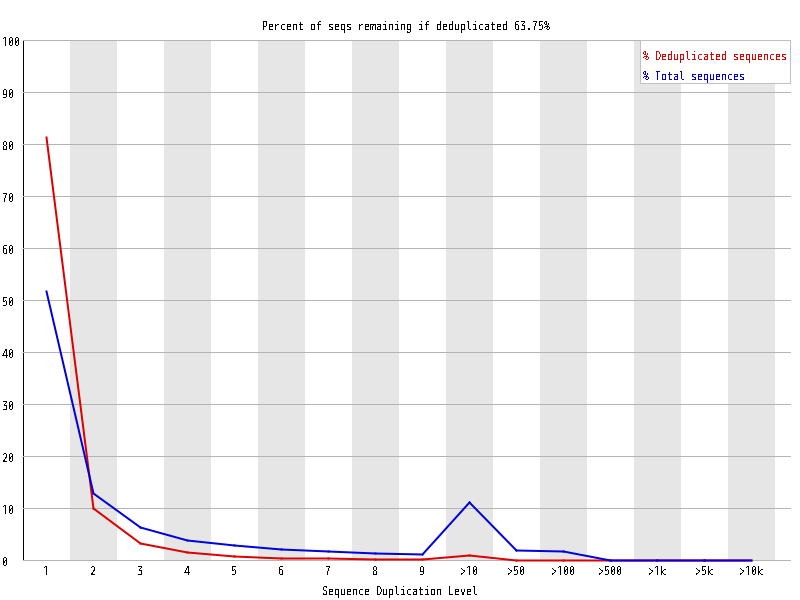
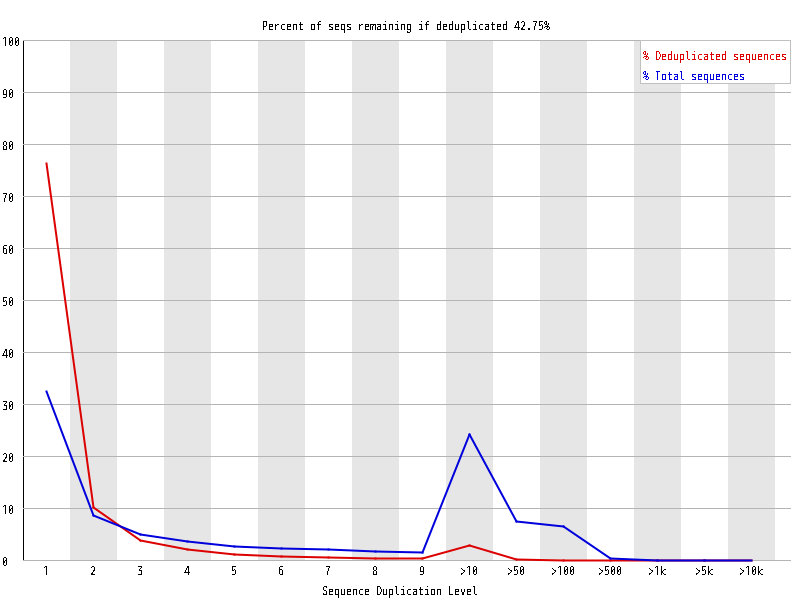
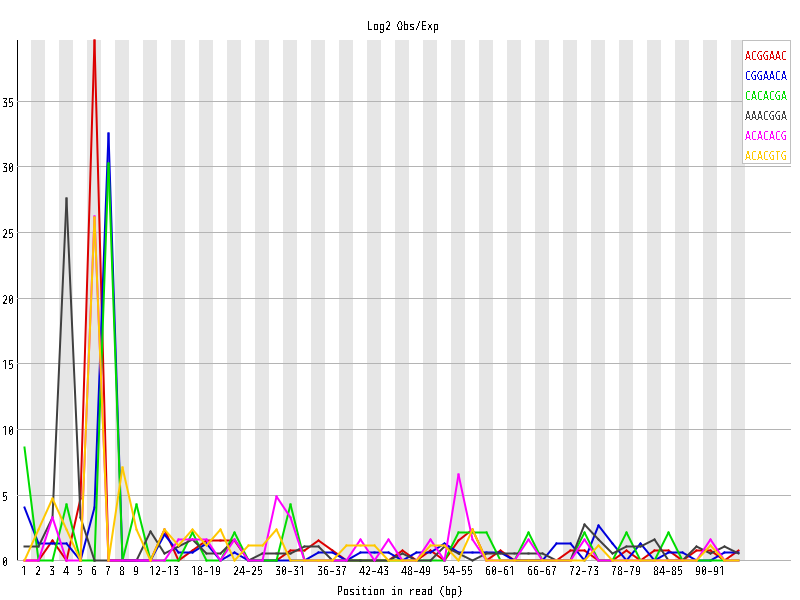
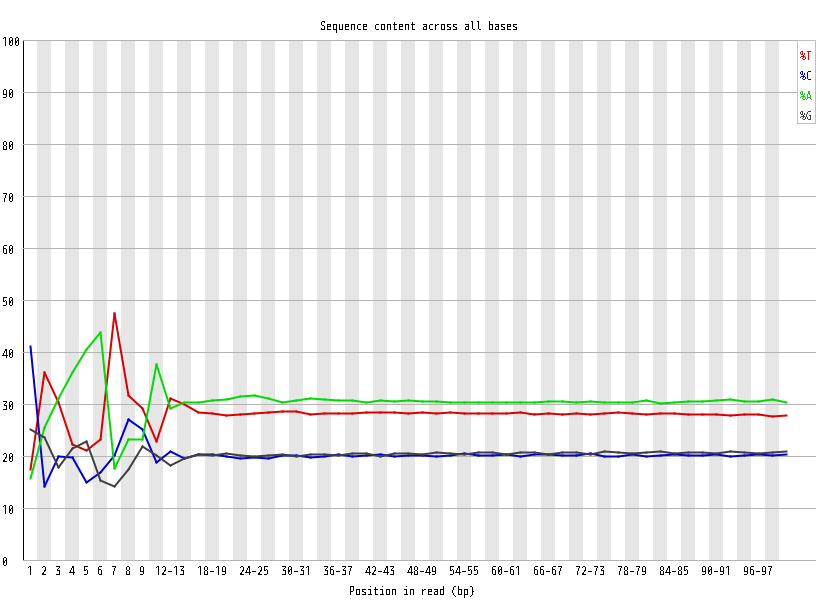
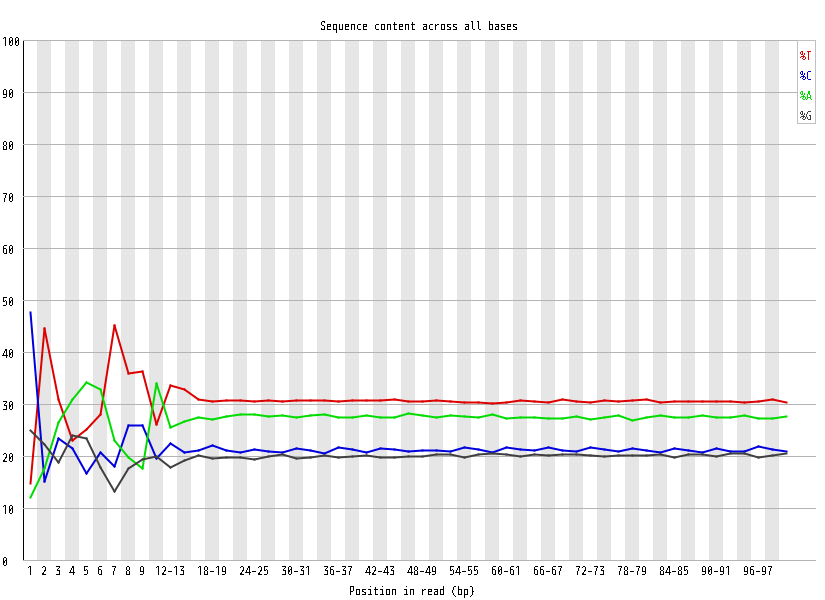
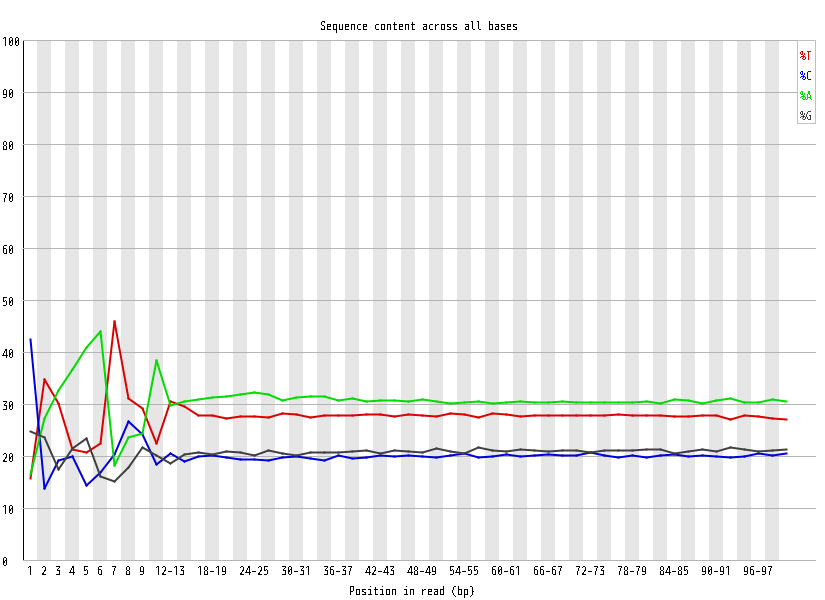
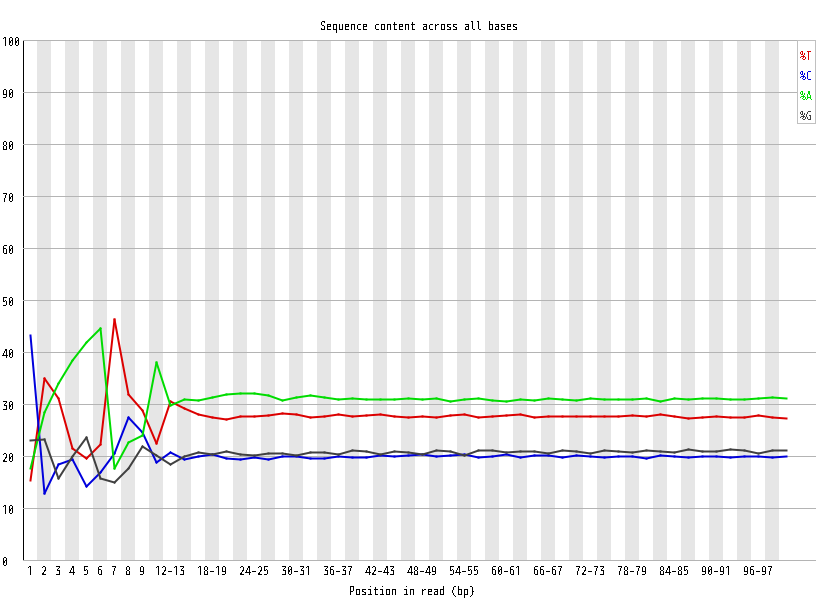
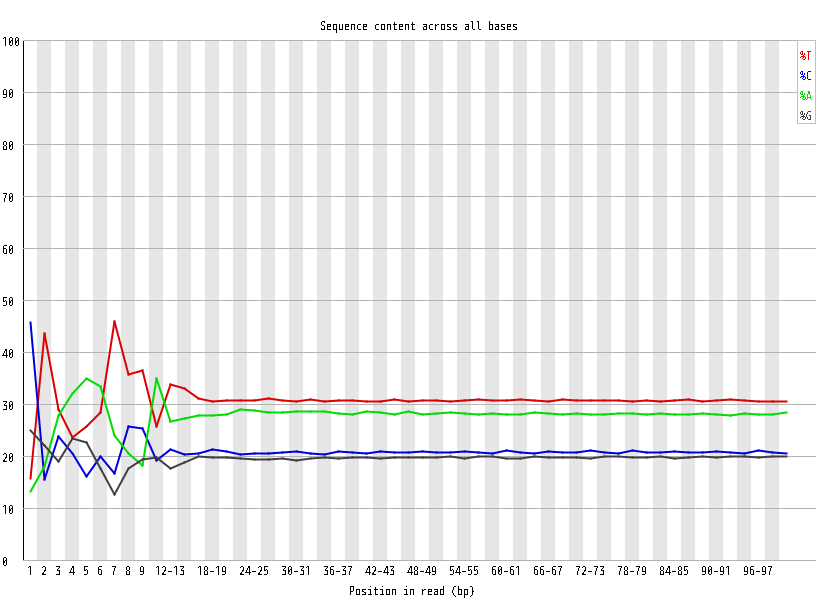
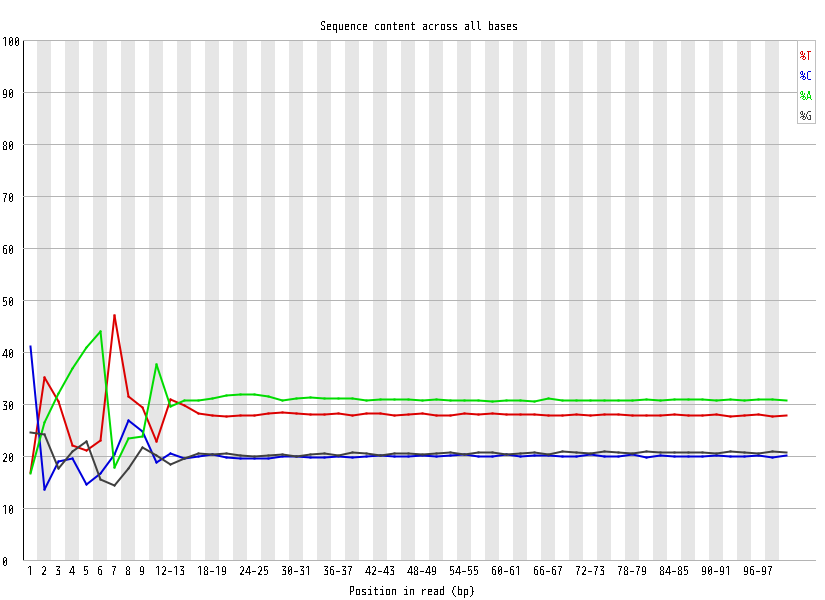
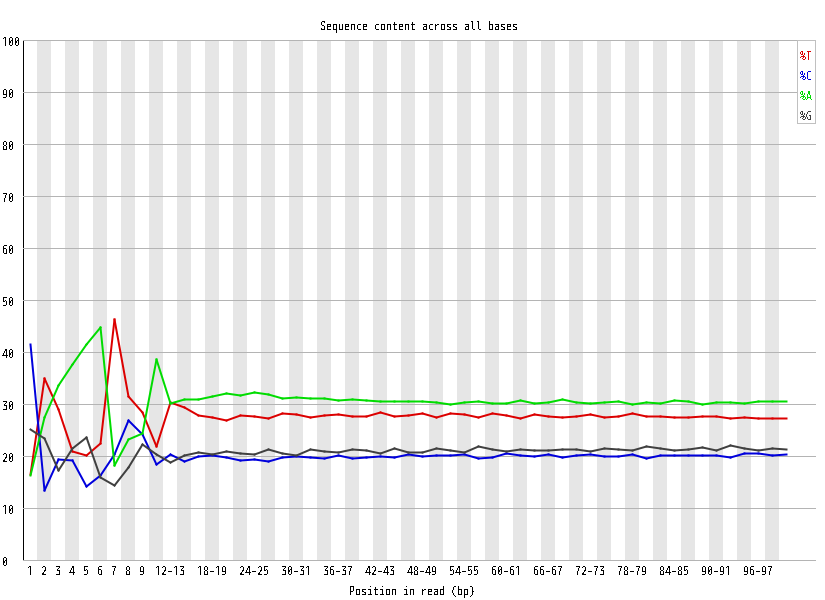
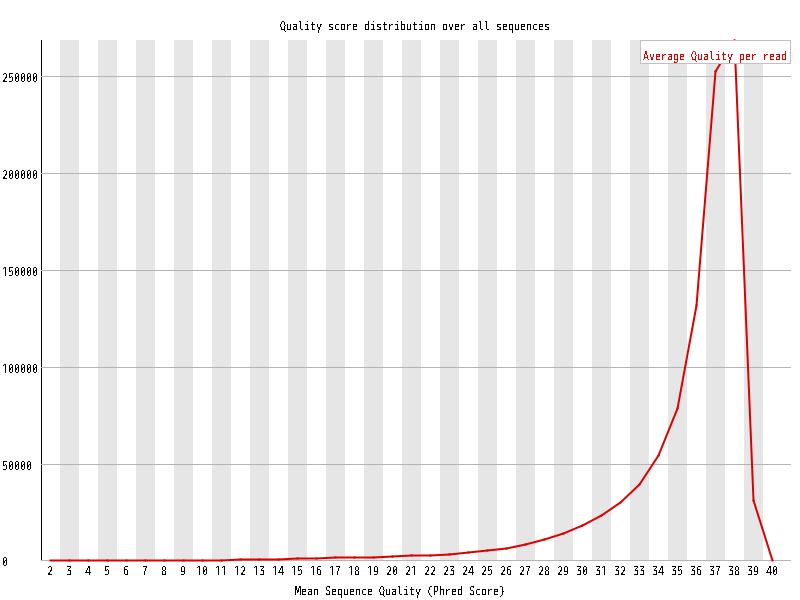
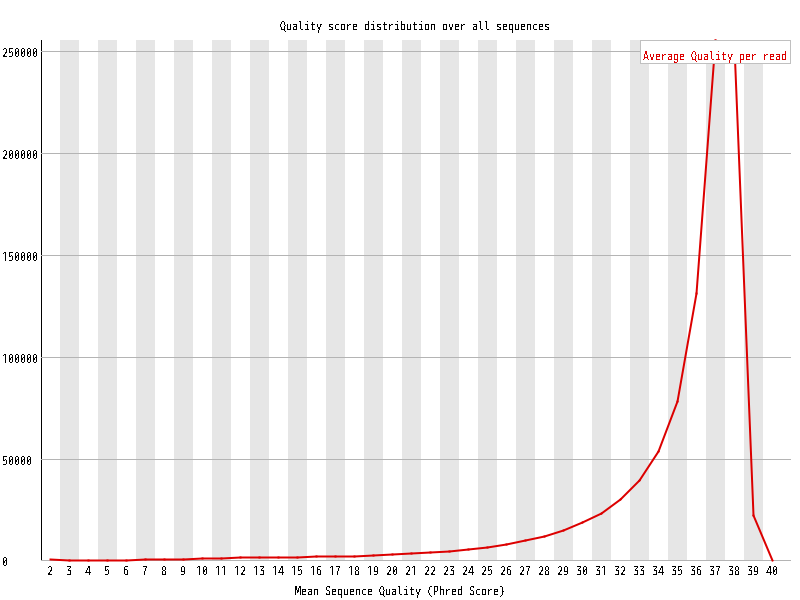
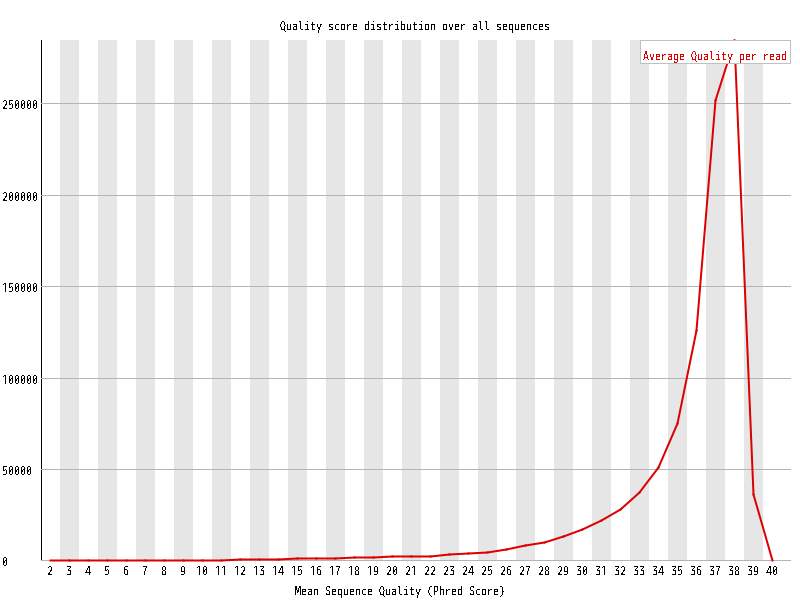
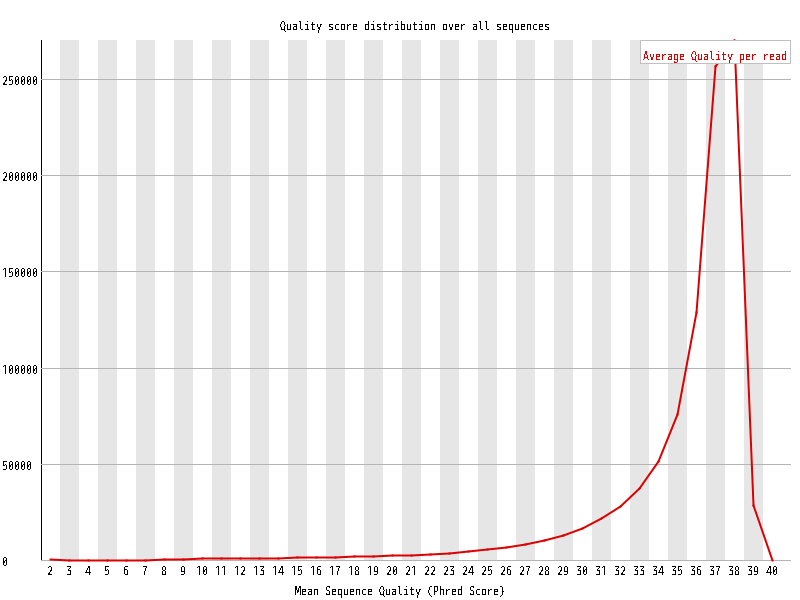
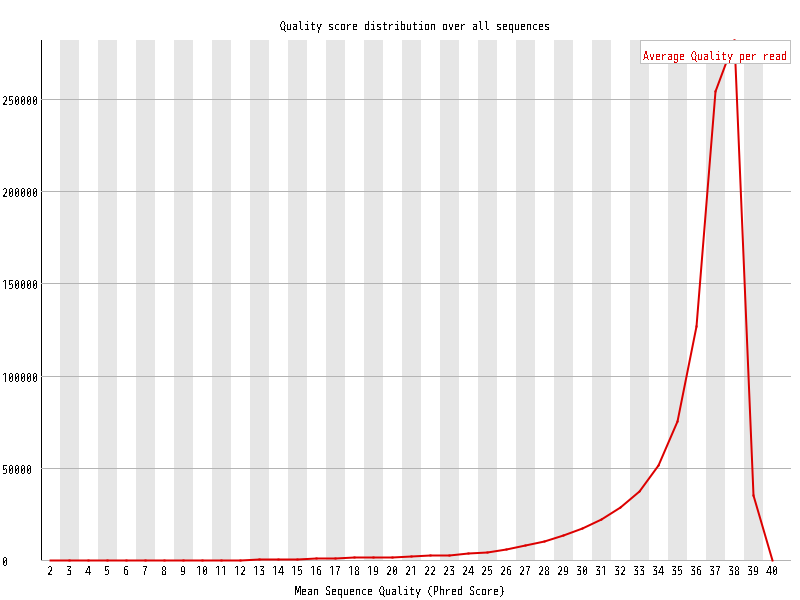
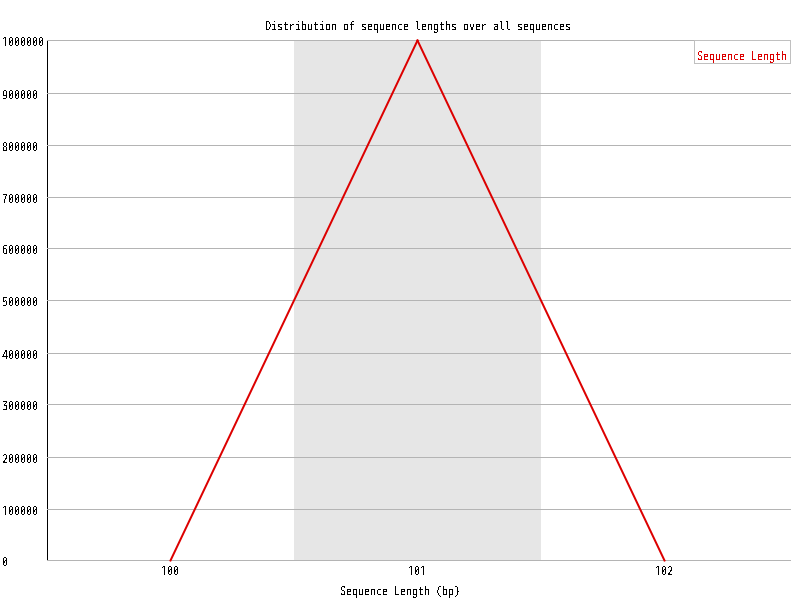
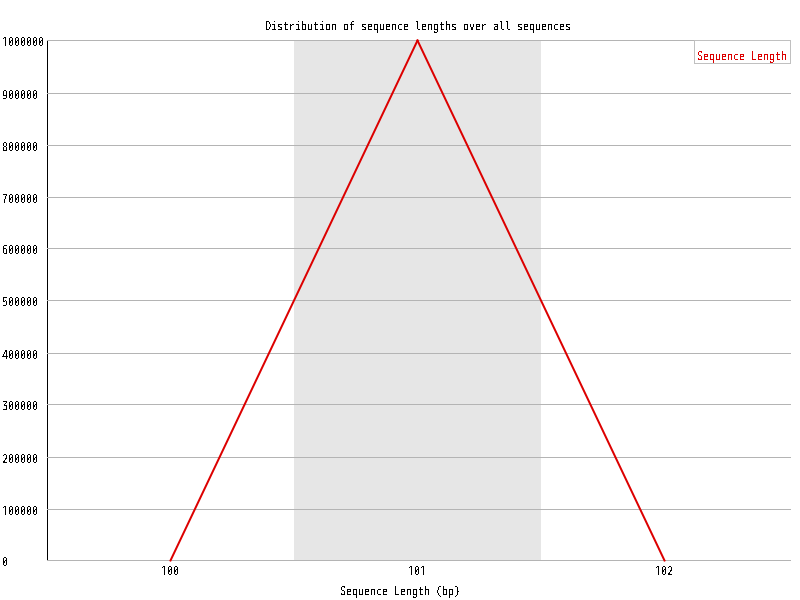
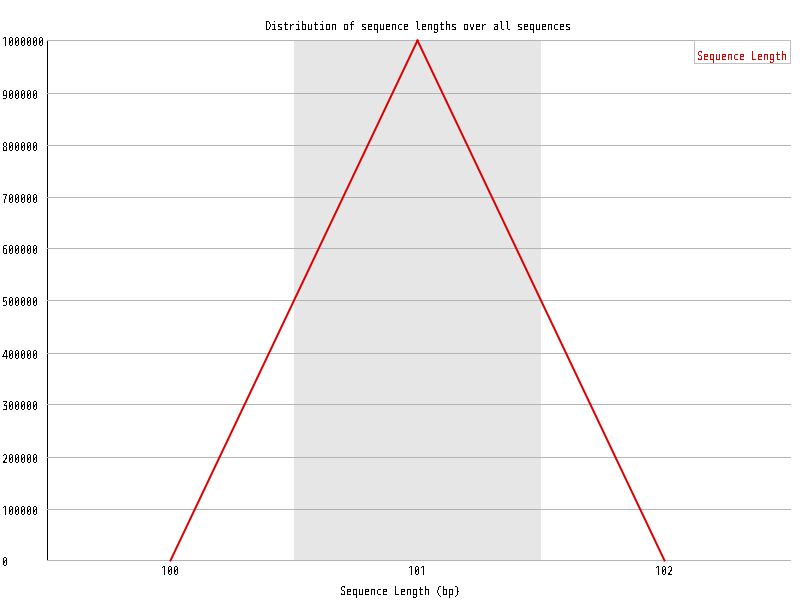
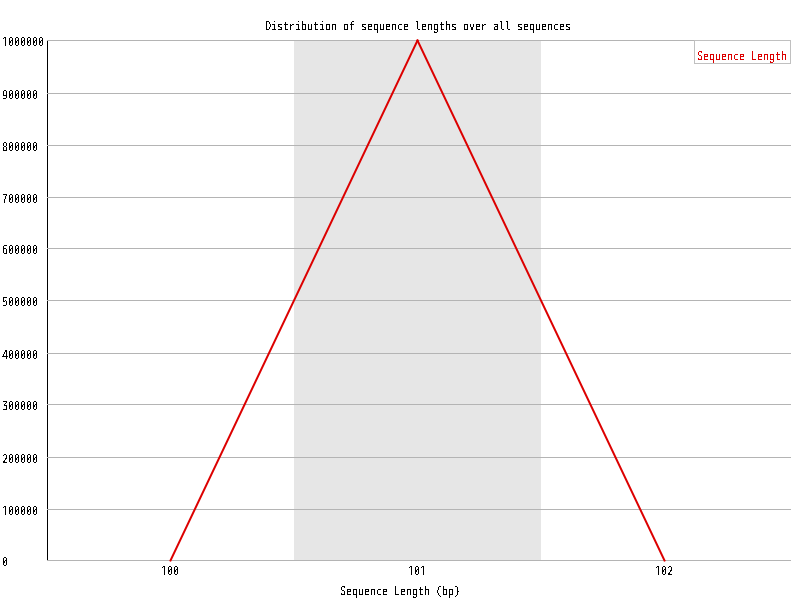
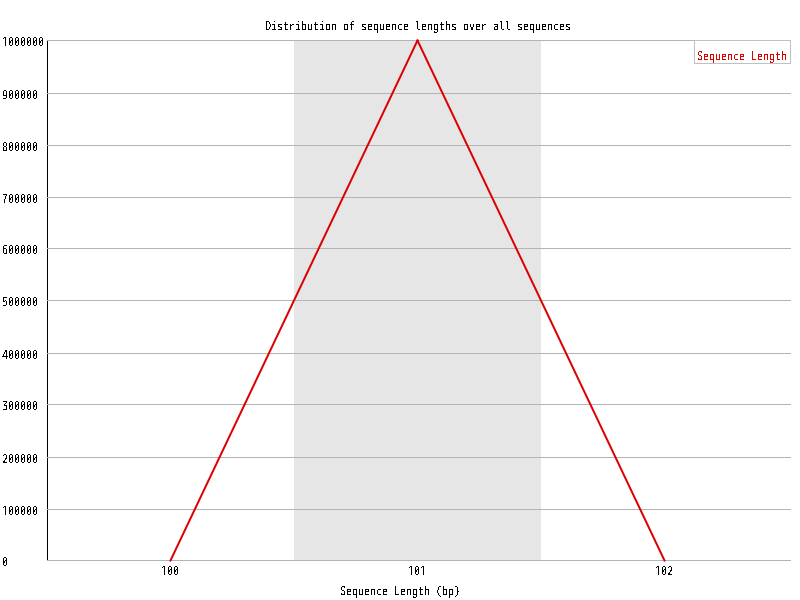
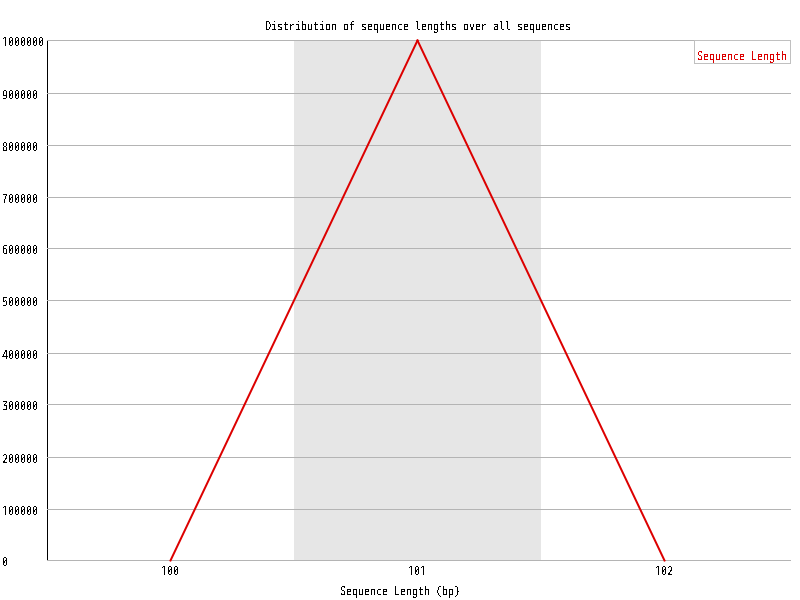
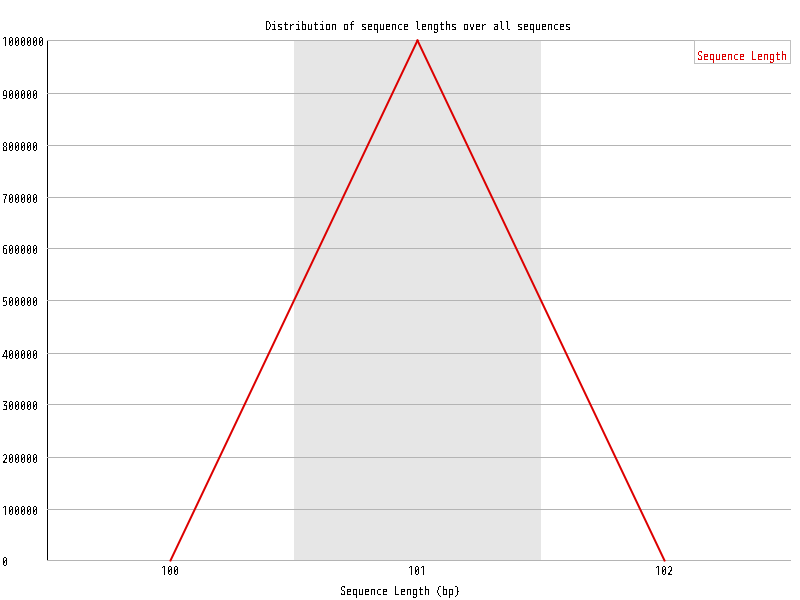
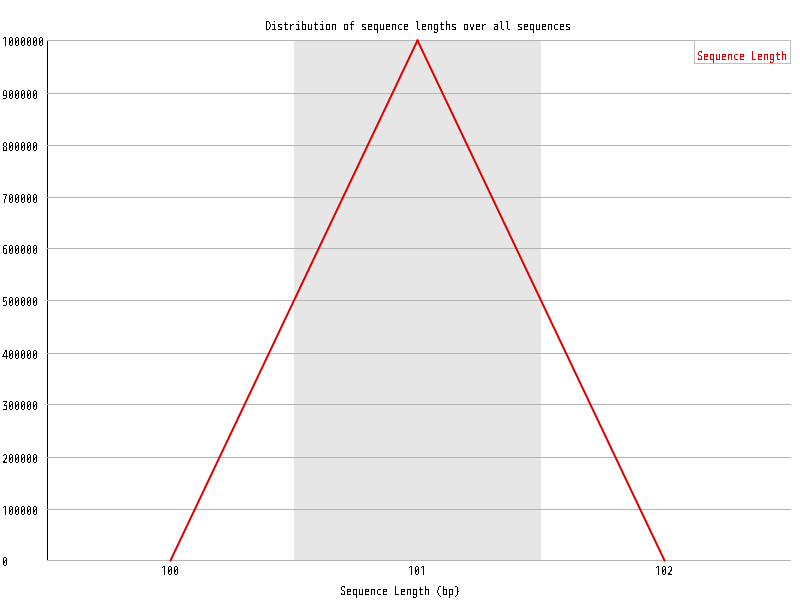
| Overrepresented sequences |
|
CTCACATTAACGTTGTCGTTATCGGTCATGTCGATTCTGGTAAGTCTACC 1344 0.1344 No Hit
CAATAATGCAATTTTCTACTGTCGCTTCTATCGCCGCTGTCGCCGCTGTC 1213 0.1213 No Hit
|
|
|
|
CAAGAAACAAAAATTCAAGTAAATAACAGATAATATGTCAAAAGCTGTCG 1252 0.1252 No Hit
CAAAAATTCAAGTAAATAACAGATAATATGTCAAAAGCTGTCGGTATTGA 1202 0.1202 No Hit
CTCTATTAAAACAGGTATCCAAAAAAGCAAACAAACAAACTAAACAAATT 1184 0.11839999999999999 No Hit
CAAAAACAAGAAAAAAAATAAACAAAACAATAATCATGTCAAAAGCTGTT 1151 0.1151 No Hit
CAGATCTCTATTAAAACAGGTATCCAAAAAAGCAAACAAACAAACTAAAC 1003 0.1003 No Hit
|
|
|
|
|
|
CGAAAACAAAAGAAAAAATAAATAAAGAAATAATCATGTCAAAAGCTGTT 2121 0.2121 No Hit
CAACAACCAAGAGGAAAAGTAACTAATAAATCCTATTAAAATAGAGAATG 1091 0.1091 No Hit
GGAAAAGTAACTAATAAATCCTATTAAAATAGAGAATGTTTTCTATATTC 1026 0.1026 No Hit
CAAGAATTCAAATAATTAATAATATAATATGTCAAAAGCTGTCGGTATTG 1014 0.10139999999999999 No Hit
|